Step 1: fitting the null logistic/linear mixed model
- When a sparse GRM is used to fit the null model (–useSparseGRMtoFitNULL=TRUE)
- Multiple variance ratios need to be estimated based on different minor allele count categories with –isCateVarianceRatio=TRUE ** Different from Step 1 in SAIGE for single-variant tests in SAIGE, in which only a single variance ratio is estiamted ** By default, two variance ratios are estiamted for 10 <= MAC < 20 and MAC >= 20. ** Use –cateVarRatioMinMACVecExclude and –cateVarRatioMaxMACVecInclude to modify the MAC categories ** Note that the PLINK file need to contain at least 200 variants whose MAC fall in these categories. ** This PLINK file is ONLY used for variance ratio estimation and contains random markers extracted from the PLINK file that is LD pruned and used for GRM and variance ratio estimation
```
#(Optional) get ids for 1000 random markers for each MAC category
## calcuate allele counts for each marker in the large plink file
plink2 --bfile ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr --freq count --out ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr
## randomly extract IDs for markers falling in the two MAC categories
cat <(tail -n +2 ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr.acount | awk '((2*$6-$5) < 20 && (2*$6-$5) >= 10) || ($5 < 20 && $5 >= 10) {print $2}' | shuf -n 1000) \
<(tail -n +2 ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr.acount | awk ' $5 >= 20 && (2*$6-$5)>= 20 {print $2}' | shuf -n 1000) > ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr.forCate_vr.markerid.list
## extract markers from the large plink file
plink2 --bfile ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr --extract ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr.forCate_vr.markerid.list --make-bed --out ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr.forCate_vr
#run step 1
Rscript step1_fitNULLGLMM.R \
--sparseGRMFile=output/sparseGRM_relatednessCutoff_0.125_1000_randomMarkersUsed.sparseGRM.mtx \
--sparseGRMSampleIDFile=output/sparseGRM_relatednessCutoff_0.125_1000_randomMarkersUsed.sparseGRM.mtx.sampleIDs.txt \
--plinkFile=./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr.forCate_vr \
--useSparseGRMtoFitNULL=TRUE \
--phenoFile=./input/pheno_1000samples.txt_withdosages_withBothTraitTypes.txt \
--phenoCol=y_binary \
--covarColList=x1,x2 \
--qCovarColList=x2 \
--sampleIDColinphenoFile=IID \
--traitType=binary \
--isCateVarianceRatio=TRUE \
--outputPrefix=./output/example_binary_sparseGRM \
--IsOverwriteVarianceRatioFile=TRUE
```
-
When a full GRM is used to fit the null model (GRM is constructed on-the-fly using genotypes in the PLINK file, –plinkfile=)
- Use a full GRM to fit the null model(by default, –useSparseGRMtoFitNULL=FALSE) with multiple CPUs ( –nThreads)
- Use both a full GRM (to be constrcuted using genotypes in the PLINK file) and a sparse GRM with –useSparseGRMforVarRatio=TRUE to estimate the variance ratio(s) ** Use –sparseGRMFile for the file containing the sparse GRM ** Use –sparseGRMSampleIDFile for the file containing the IDs for samples in the sparse GRM
- Multiple variance ratios need to be estimated based on different minor allele count categories with –isCateVarianceRatio=TRUE ** By default, two variance ratios are estiamted for 10 <= MAC < 20 and MAC >= 20. ** Use –cateVarRatioMinMACVecExclude and –cateVarRatioMaxMACVecInclude to modify the MAC categories ** Note that the PLINK file need to contain at least 200 variants whose MAC fall in these categories ** Different from Step 1 in SAIGE for single-variant tests in SAIGE, in which only a single variance ratio is estiamted
Rscript step1_fitNULLGLMM.R \ --plinkFile=./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly_22chr \ --sparseGRMFile=output/sparseGRM_relatednessCutoff_0.125_1000_randomMarkersUsed.sparseGRM.mtx \ --sparseGRMSampleIDFile=output/sparseGRM_relatednessCutoff_0.125_1000_randomMarkersUsed.sparseGRM.mtx.sampleIDs.txt \ --phenoFile=./input/pheno_1000samples.txt_withdosages_withBothTraitTypes.txt \ --phenoCol=y_binary \ --covarColList=x1,x2 \ --qCovarColList=x2 \ --sampleIDColinphenoFile=IID \ --traitType=binary \ --outputPrefix=./output/example_binary_fullGRM \ --nThreads=64 \ --useSparseGRMtoFitNULL=FALSE \ --isCateVarianceRatio=TRUE \ --useSparseGRMforVarRatio=TRUE \ --IsOverwriteVarianceRatioFile=TRUE
Input files (Same as input in SAIGE Step 1)
- Same as the input in Step 1 by SAIGE for single-variant association tests
- Specific to SAIGE-GENE+ for rare variants, the PLINK file –plinkFile= needs to contain rare variants with MAC low to 10.
- (Required) Phenotype file (contains covariates if any, such as gender and age) The file can be either space or tab-delimited with a header. It is required that the file contains one column for sample IDs and one column for the phenotype. It may contain columns for covariates.
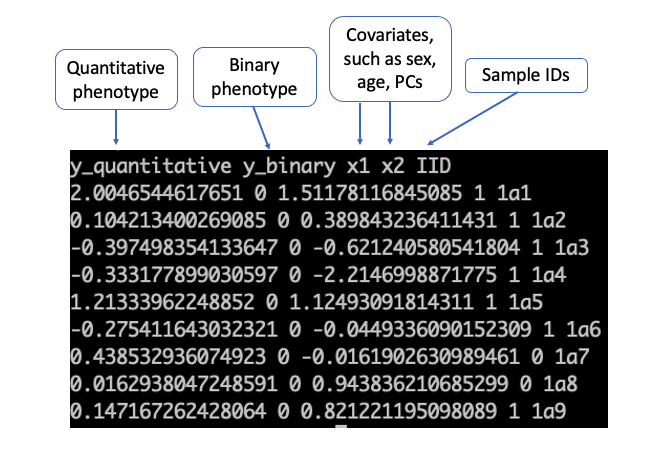
```
less -S ./input/pheno_1000samples.txt_withdosages_withBothTraitTypes.txt
```
-
(Optional) Genotype file for constructing the full genetic relationship matrix (GRM) and estimating the variance ratio
SAIGE takes the PLINK binary file for the genotypes and assumes the file prefix is the same one for .bed, .bim. and .fam./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly.bed ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly.bim ./input/nfam_100_nindep_0_step1_includeMoreRareVariants_poly.fam -
(Optional) sparse GRM file and the sample ID file for the sparse GRM. Please see Step 0 for more details.
./output/sparseGRM_relatednessCutoff_0.125_1000_randomMarkersUsed.sparseGRM.mtx ./output/sparseGRM_relatednessCutoff_0.125_1000_randomMarkersUsed.sparseGRM.mtx.sampleIDs.txt ##the sparse matrix can be read and viewed using the R library Matrix library(Matrix) sparseGRM = readMM("./output/sparseGRM_relatednessCutoff_0.125_1000_randomMarkersUsed.sparseGRM.mtx")
Output files ((Same as output in SAIGE Step 1 for single-variant association tests))
- Same as the output in Step 1 by SAIGE for single-variant association tests
- Specific to SAIGE-GENE+ for rare variants, the variance ratio file contains multiple variance ratios instead a single variance ratio
-
model file (input for Step 2)
./output/example_binary.rda #load the model file in R load("./output/example_binary.rda") names(modglmm) modglmm$theta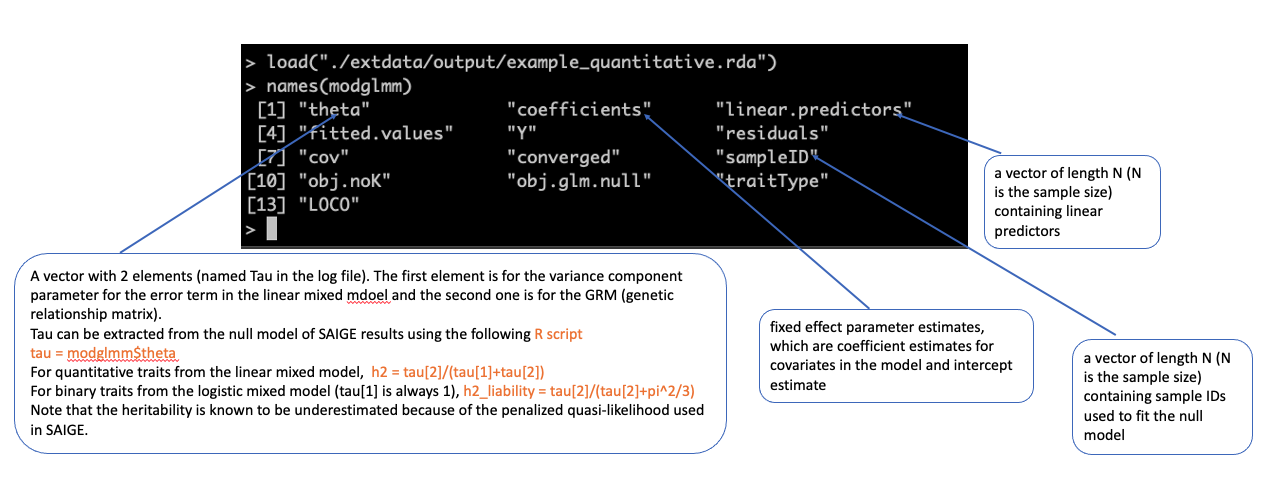
-
variance ratio file (if categorical variance ratios are estiamted in Step 1, this will be input for Step 2)
less -S ./output/example_binary_cate.varianceRatio.txt